Introduction
Recently I read an article in Coding the Past about using ChatGPT to wrangle Wikipedia tables into a format usable in R. I tried the method, but found it didn’t work well with tables that had blank cells. Many wikipedia tables have blank cells, so this is likely to be a common issue. Following a post on pipe dreams, I turned instead to using rvest for scraping wikipedia table data. In my experience, scraping the data is a much better option than messing around with ChatGPT.
I selected the list of genocides as the table to retrieve.
Select the table and scrape the object
url <- "https://en.wikipedia.org/wiki/List_of_genocides"url_bow <- polite::bow(url)Inspect the page and look for HTML called table.wikitable.
ind_html <-
polite::scrape(url_bow) |> # scrape web page
rvest::html_nodes("table.wikitable") |> # pull out specific table
rvest::html_table(fill = TRUE) The rvest function returns a list of one item, so we need to index the first item in the list to get the data frame. We do so using the double square bracket [[]] index operator which returns an object of the class of item that is contained in the list.
ind_html[[1]] |>
head() | Event | Location | Period | Period | Estimated killings | Estimated killings | Proportion of group killed |
|---|---|---|---|---|---|---|
| Event | Location | From | To | Lowest | Highest | Proportion of group killed |
| Rohingya genocide[N 1] | Rakhine StateMyanmar | 2016 | Present | 9,000–13,700[9] | 43,000[10] | Before the 2015 Rohingya refugee crisis and the military crackdown in 2016 and 2017, the Rohingya population in Myanmar was around 1.0 to 1.3 million, chiefly in the northern Rakhine townships, which were 80–98% Rohingya. Since 2015, over 900,000 Rohingya refugees have fled to south-eastern Bangladesh alone, and more to other surrounding countries, and major Muslim nations. More than 100,000 Rohingyas in Myanmar are confined in camps for internally displaced persons. |
| Iraqi Turkmen genocide[N 2] | Islamic State-controlled territory in northern Iraq | 2014 | 2017 | 3,500 | 8,400 | |
| Genocide of Yazidis by the Islamic State[N 3] | Islamic State-controlled territory in northern Iraq and Syria | 2014 | 2019 | 2,100[18] | 5,000[19] | |
| Darfur genocide[N 4] | Darfur, Sudan | 2003 | Present | 98,000[22] | 500,000[23] | |
| Effacer le tableau[N 5] | North Kivu, Democratic Republic of the Congo | 2002 | 2003 | 60,000[26][24] | 70,000[26] | 40% of the Eastern Congo’s Pygmy population killed[N 6] |
Remove footnotes and commas and convert strings into numbers
This worked well, but there are still lingering characters from footnotes. We want to remove the brackets and the numbers inside them, but leave the rest of the text. These can be removed with regular expressions…so time to turn to StackOverflow. We’ll start with an isolated example, ensure that works properly, and then attempt to apply it to the entire data frame.
test <- "2,100[17]"To do this, we’ll need to use the double escape //.
test2 <- gsub("\\[([^]]+)\\]", "",test)
test2[1] "2,100"lapply(ind_html[[1]], function(x) gsub("\\[([^]]+)\\]", "",x)) |>
as.data.frame() |>
head()| Event | Location | Period | Period.1 | Estimated.killings | Estimated.killings.1 | Proportion.of.group.killed |
|---|---|---|---|---|---|---|
| Event | Location | From | To | Lowest | Highest | Proportion of group killed |
| Rohingya genocide | Rakhine StateMyanmar | 2016 | Present | 9,000–13,700 | 43,000 | Before the 2015 Rohingya refugee crisis and the military crackdown in 2016 and 2017, the Rohingya population in Myanmar was around 1.0 to 1.3 million, chiefly in the northern Rakhine townships, which were 80–98% Rohingya. Since 2015, over 900,000 Rohingya refugees have fled to south-eastern Bangladesh alone, and more to other surrounding countries, and major Muslim nations. More than 100,000 Rohingyas in Myanmar are confined in camps for internally displaced persons. |
| Iraqi Turkmen genocide | Islamic State-controlled territory in northern Iraq | 2014 | 2017 | 3,500 | 8,400 | |
| Genocide of Yazidis by the Islamic State | Islamic State-controlled territory in northern Iraq and Syria | 2014 | 2019 | 2,100 | 5,000 | |
| Darfur genocide | Darfur, Sudan | 2003 | Present | 98,000 | 500,000 | |
| Effacer le tableau | North Kivu, Democratic Republic of the Congo | 2002 | 2003 | 60,000 | 70,000 | 40% of the Eastern Congo’s Pygmy population killed |
So far so good, the footnote characters are gone, so lets save this as a dataframe. However, there is a bit more work to do before this can be graphed.
df <- lapply(ind_html[[1]], function(x) gsub("\\[([^]]+)\\]", "",x)) |>
as.data.frame() Strings that contain a comma are coerced to NA and this isn’t useful. So the commas need to be removed
as.numeric(test2)[1] NAWe can use gsub() to remove the , and then convert to numeric.
gsub(",", "", test2) |>
as.numeric()[1] 2100We need to remove the commas from any column that has numbers, but we want to keep commas in columns that are text. The low end estimate column has some ranges that are separated by a - so we would want to either take the numbers before or after this character. Since these are supposed to be low estimates, we’ll take numbers before and remove everything from the - to the end.
df$Estimated.killings <- as.numeric(gsub(",", "", df$Estimated.killings))sample(df$Estimated.killings, 20) [1] 200000 248000 130000 2100 10000 480000 491000 80000 300000
[10] 34000 3500 50000 600000 111091 50000 60000 34000 83000
[19] 4204000 NAPlotting the table: sorted bar chart and long x values
We can plot loss of life by event using a bar chart.
The plot presents two fairly common challenges.
Some of the label text is very long. As described in this post and the documentation, we can use the
scale_x_discrete()function with thelabel_wrap_gen()argument.The numbers are very large and would be better represented as an abbreviation. This can be achieved with the
scales()package using thelabel_number()argumentscale_cutwith a value ofcut_short_scale(). The documentation and a tidyverse blog post provide some other options.
df |>
dplyr::filter(!is.na(Estimated.killings)) |> # remove NA values
ggplot(aes(x = reorder(Event,Estimated.killings), y = Estimated.killings)) +
geom_bar(stat = "identity", position=position_dodge(.5)) +
scale_x_discrete(labels = label_wrap_gen(50))+
scale_y_continuous(labels = scales::label_number(scale_cut = scales::cut_short_scale())) +
coord_flip()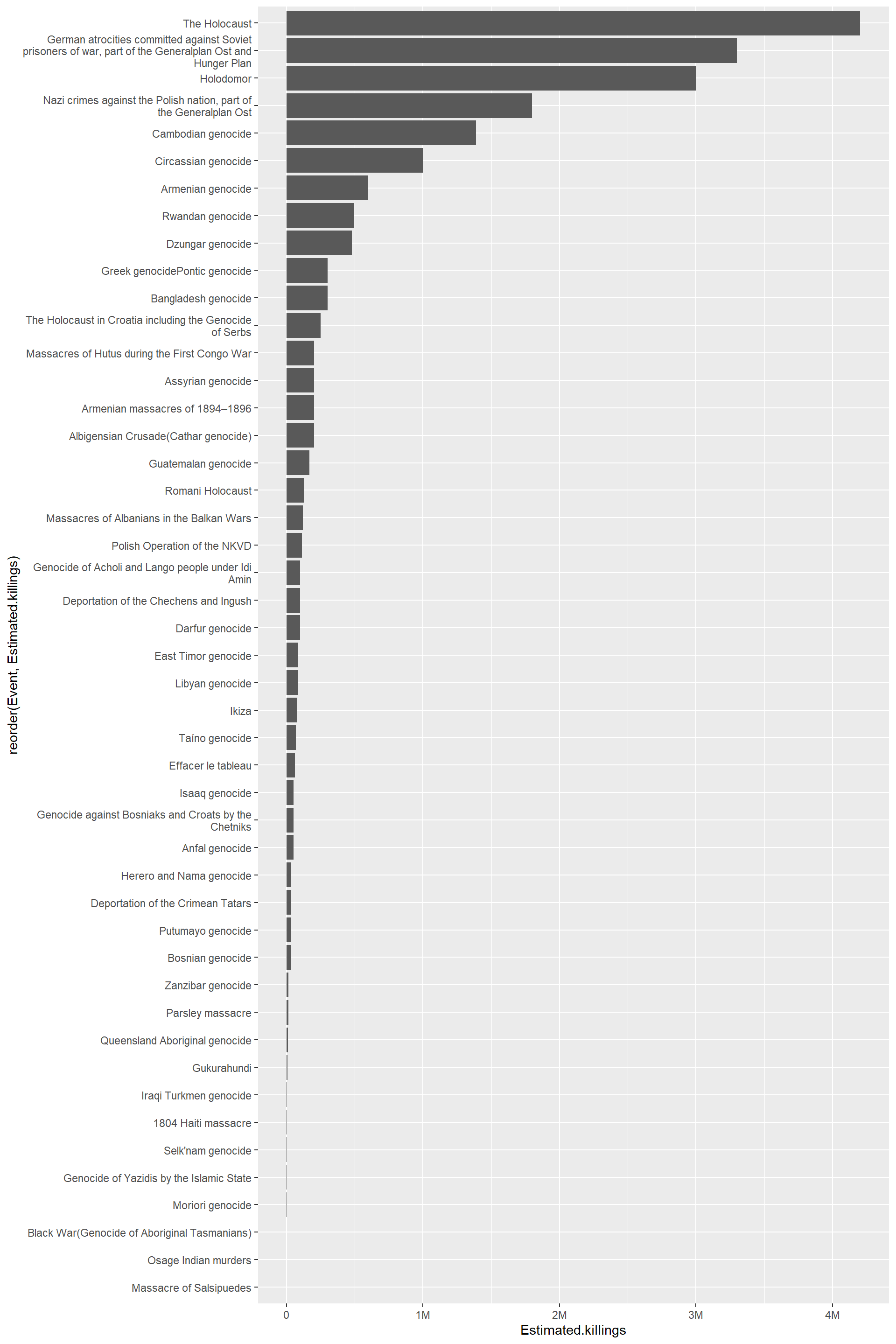
Just noting that if we reorder based on location, the reorder function does not work properly (I did this). The issue is that there three locations where more than one genocide is listed on the table.
| Location | n | |
|---|---|---|
| 42 | Ukraine and the heavily Ukrainian-populated northern Kuban, in the Soviet Union | 1 |
| 43 | Uruguay | 1 |
| 44 | Van Diemen’s Land (now Australia) | 1 |
| 45 | Zanzibar (now part of Tanzania) | 1 |
| 46 | Ottoman Empire (now Turkey, Syria and Iraq) | 2 |
| 47 | German-occupied Europe | 3 |
According to this answer over at the RStudio forums, ggplot sums the values within each of these locations. In the case of German-occupied Europe, there are three genocides summed so the value is exceptionally large. However, Ottoman Empire (now Turkey, Syria and Iraq) is also not displaying properly. It just isn’t as easy to spot.
df |>
dplyr::filter(!is.na(Estimated.killings)) |> # remove NA values
ggplot(aes(x = reorder(Location,Estimated.killings), y = Estimated.killings)) +
geom_bar(stat = "identity") +
scale_y_continuous(labels = scales::label_number(scale_cut = scales::cut_short_scale()))+
scale_x_discrete(labels = label_wrap_gen(40))+
coord_flip()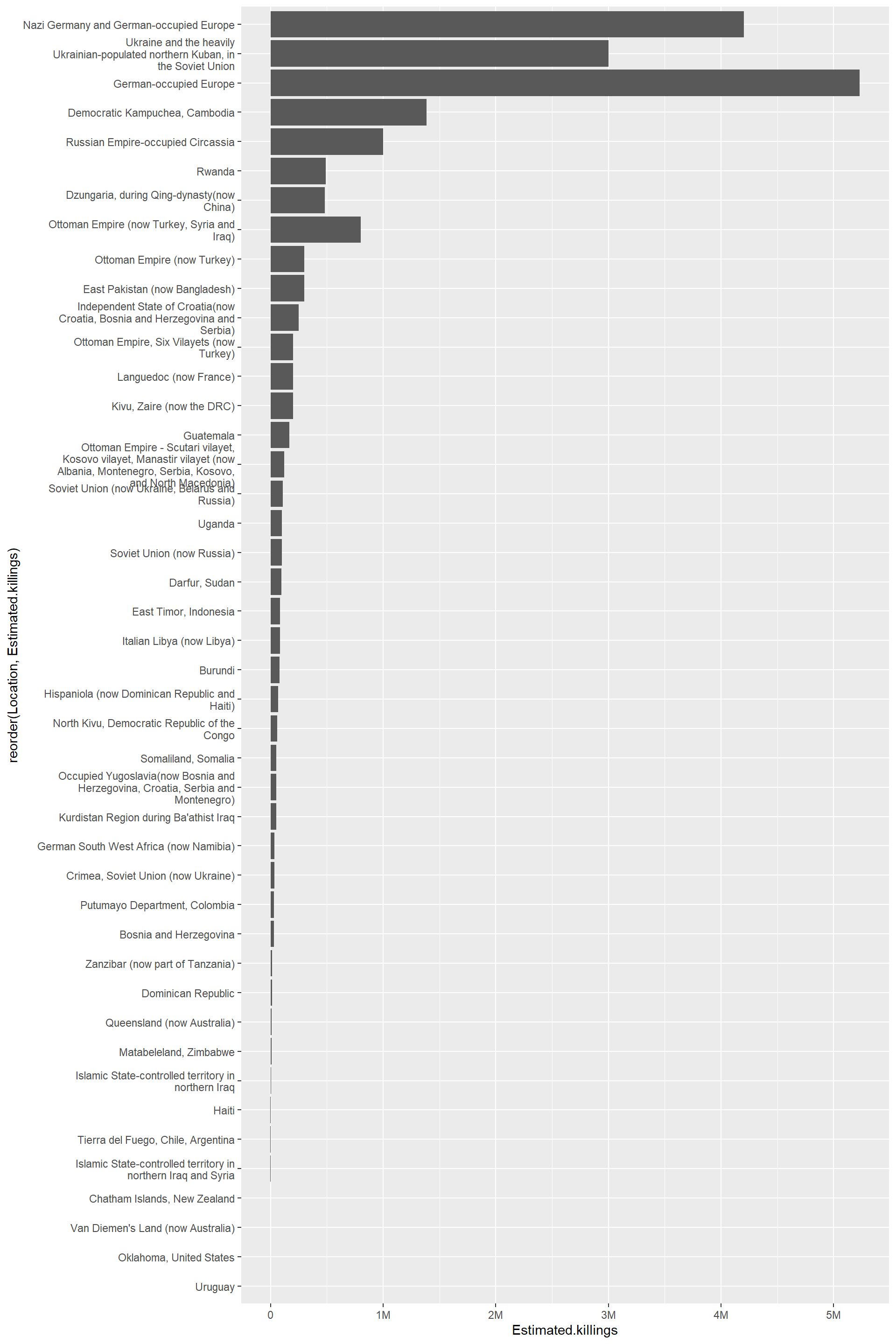
- python - How can I remove text within parentheses with a regex? - Stack Overflow
- regex - Regular expression to extract text between square brackets - Stack Overflow
- r - How do I deal with special characters like ^$.?*|+()[{ in my regex? - Stack Overflow
- r - Applying gsub to various columns - Stack Overflow Specifically, this answer worked in the present case.
Citation
@online{craig2023,
author = {Craig, Nathan},
title = {Scraping {Wikipedia} {Table:} {Genocides}},
date = {2023-11-01},
url = {https://ncraig.netlify.app/posts/2023-11-01-wikipedia-table-scrape/index.html},
langid = {en}
}